The full phenotypic releases of the Enhanced NKI-RS Sample require a data usage agreement – a requirement similar to efforts such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and the National Database for Autism Research (NDAR). The adoption of a data usage agreement is not intended to limit the specific analyses a researcher can perform; users will only need to specify the broad range of analyses they may pursue with the data (e.g., association studies between DTI, R-fMRI, and behavior), not a specific analysis or set of analyses. The intent of the agreement is to ensure that data users agree to protect participant confidentiality when handling data that contains potentially identifying information and that they will agree to take the necessary measures to prevent breaches of privacy. The specific agreement to be employed for the Enhanced NKI-RS are those previously defined by the New York State Office of Mental Health, and can be found in the Data Usage Agreement (DUA).
Unlike the NDAR agreement, institutional review board (IRB) approval is not required for transfer of the data; it will be up to the individual data user to satisfy any additional requirements specified by their local IRB or ethics committee, prior to using the NKI-RS. Given that local IRB approval is not required as part of an individuals application for access to the NKI-RS, there is no need for an individual’s IRB to have a federal-wise assurance number – which can limit recipients of the NDAR datasets.
For your convenience, the DUA is already completed in its entirety with NKI-approved text for data handling. Investigators simply need to:
- Download the DUA form.
- Review all contents.
- Provide appropriate information regarding their name(s) and institution.
- Have the document signed and notarized by the appropriate institutional representative.
- Scan and send us the document.
- When your application has been received and approved, you will receive an email confirming your ability to request all phenotypic (single-item and summary) and imaging data in the Longitudinal Online Research and Imaging System (LORIS) Database or the Collaborative Informatics and Neuroimaging Suite (COINS) (see descriptions below).
Note: If your institution requests any clarifications regarding the DUA, just contact us and let us know – we are happy to work with them.
Accessing Phenotypical Databases
After receiving the DUA, you may access the full phenotypical data through two database portals, LORIS or COINS. Contact us if you have any issues accessing LORIS or COINS.
As of December 2019, we reorganized the NKI-RS dataset across all contributing studies to facilitate ease of data handling for either individual study or aggregate data exports within the LORIS database: Rockland Sample LORIS database. Instructions on how to access phenotypic data through LORIS is below. Note, direct links to the MRI images in the AWS S3 bucket in BIDS format can be easily obtained through LORIS.
We recommend using only one point of access (LORIS OR COINS), however, if you need to compare across exported data the coding in LORIS can be mapped to COINS exports using a crosswalk downloaded from this link.
Please note that data released from both database portals are identical, however, due to filtering differences COINS exports may show less than expected data at times.
Study Codebooks
All phenotypic data is available from either database portal (LORIS/COINS). In LORIS, variable names and short descriptions can be downloaded based on your selected export file. In COINS, a static codebook can be downloaded from the website but we recommend using the one found here, updated and formatted for ease of use. The NKI-RS Codebook includes LORIS and COINS variable names (there are some slight variations). This reference can also be used for LORIS – COINS variable mappings, if needed to reconcile COINS exports with LORIS exports.
Phenotypic Database Overview
Disclaimer:
The Nathan Kline Institute-Rockland Sample is an ongoing program of research available to you pre-publication. In order to release the widest range of data, we do minimal data cleaning and validation. We highly encourage researchers who use our data to do their own validation and quality control checks. You may contact us at RocklandSample.Enduser@nki.rfmh.org with feedback, questions, or corrections.
Accessing Phenotypic Data
After a DUA is completed, fully supported data may be accessed via a LORIS platform.
Note: There are slight differences between assessment codes in LORIS and our first generation database (COINS). The crosswalk between the codes can be downloaded here.
Loris Instructions
Step 1: Requesting Access
After completing the DUA, user accounts for LORIS can be requested though the Rockland Sample contact email: RocklandSample.Enduser@nki.rfmh.org
Step 2: Logging In
The Rockland Sample data on LORIS can be accessed at: https://data.rocklandsample.rfmh.org/. Login by entering your username and password into the box labelled “Login to LORIS”. Note, currently the “Request Account” button on the LORIS website is not functional (see step 1).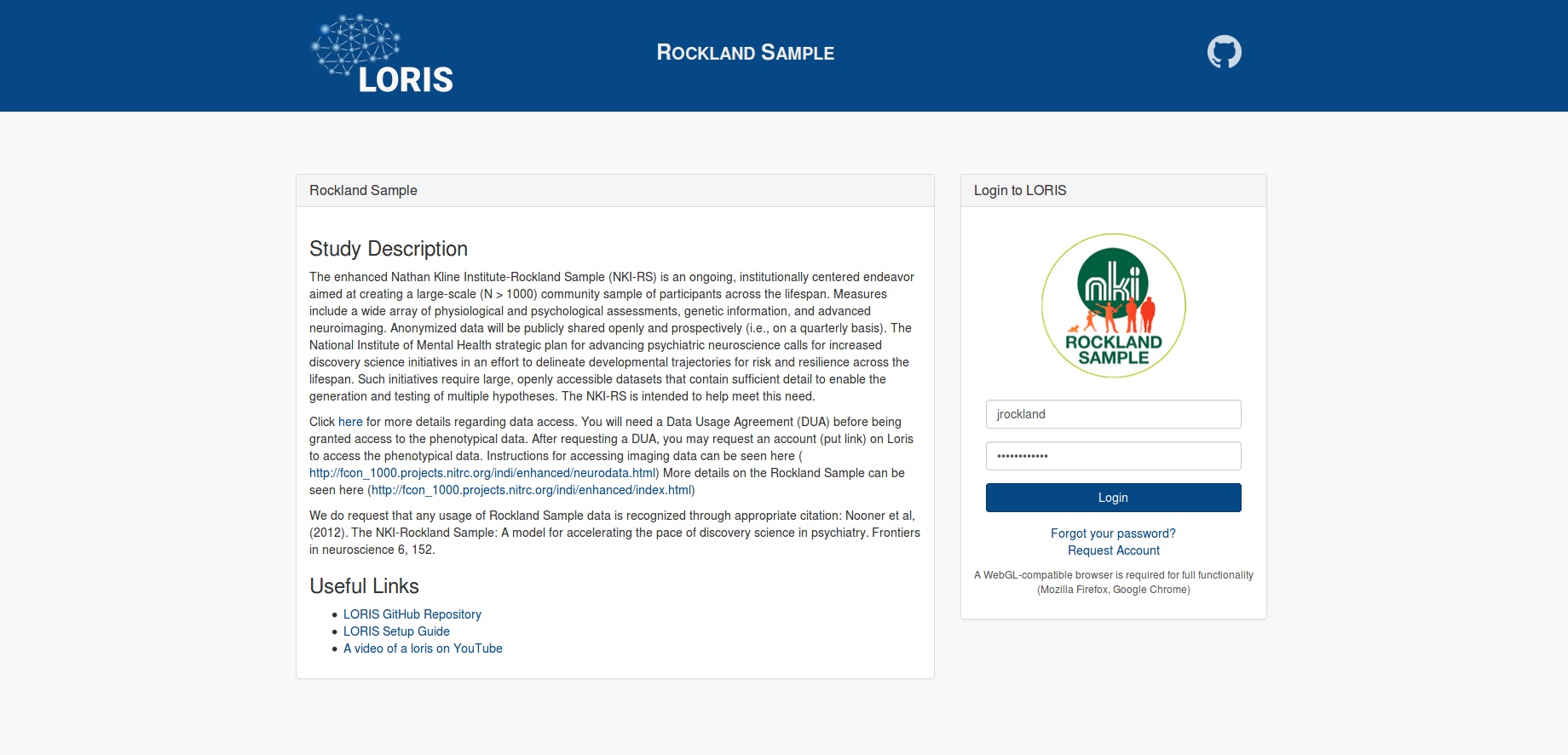 Step 3: Selecting Data
Step 3: Selecting Data
To get to the Data Query tool, go to the top bar, and select “Reports” then “Data Query Tool”.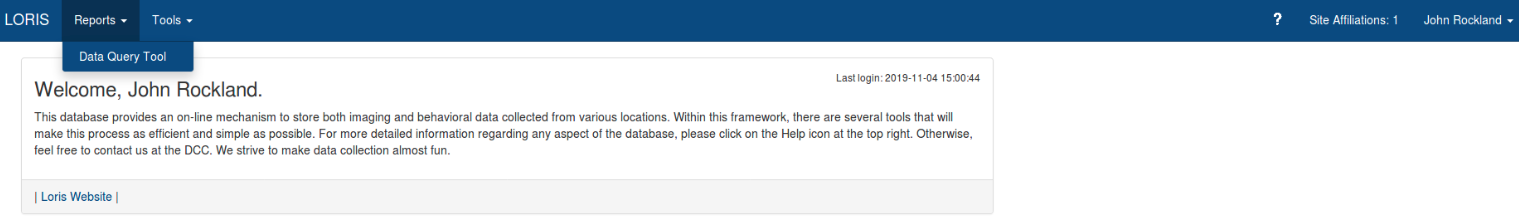
Select the tab “Define Fields”, and enter an Assessment (in LORIS they are referred as “Instrument”) that you want to include in your data export. The variables (in LORIS they are referred as “Fields”) from the instrument will populate the page.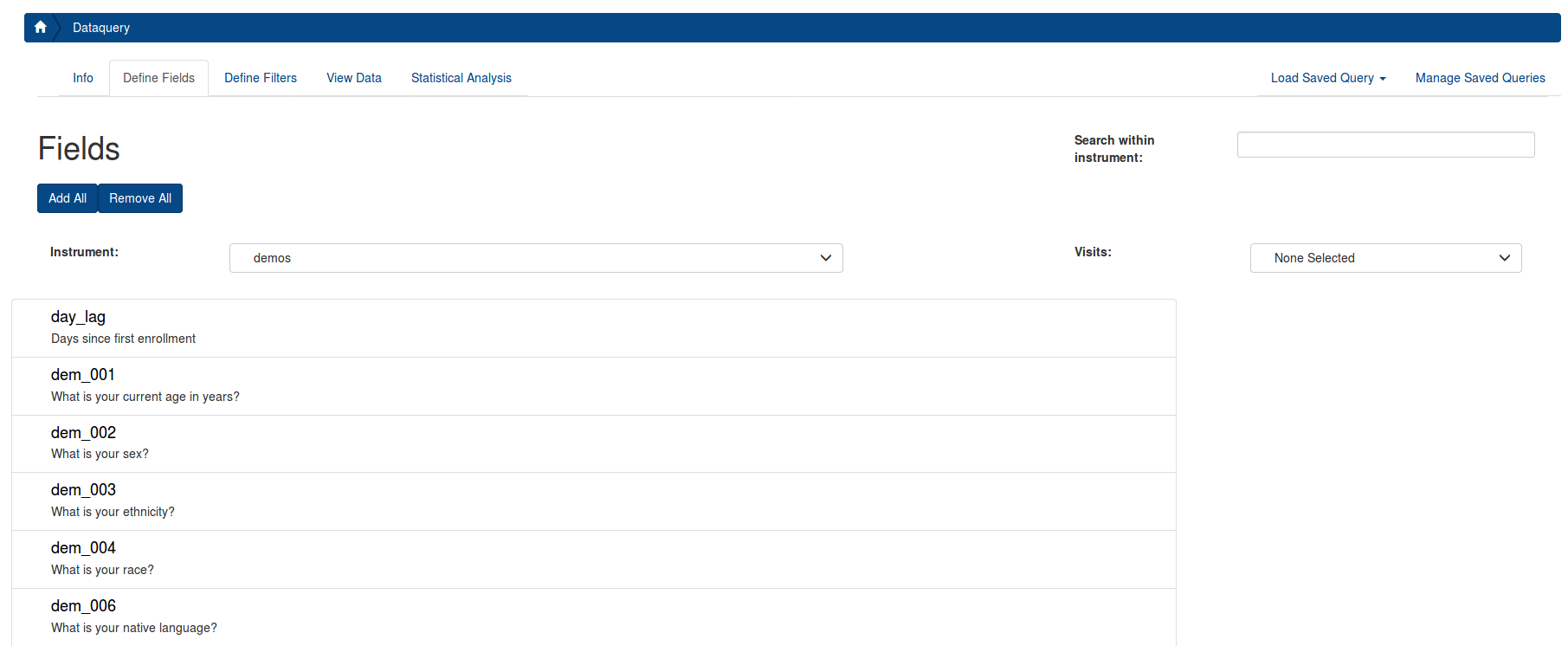 After you select your first Field (A), select Visits (B) from the box on the right (Details describing the visits can be seen here). Then, select the desired Fields by clicking the on each one (C). Selected Fields will populate on the rightmost column (D). You can scroll through all instruments to choose Fields for your exported datafile.
After you select your first Field (A), select Visits (B) from the box on the right (Details describing the visits can be seen here). Then, select the desired Fields by clicking the on each one (C). Selected Fields will populate on the rightmost column (D). You can scroll through all instruments to choose Fields for your exported datafile.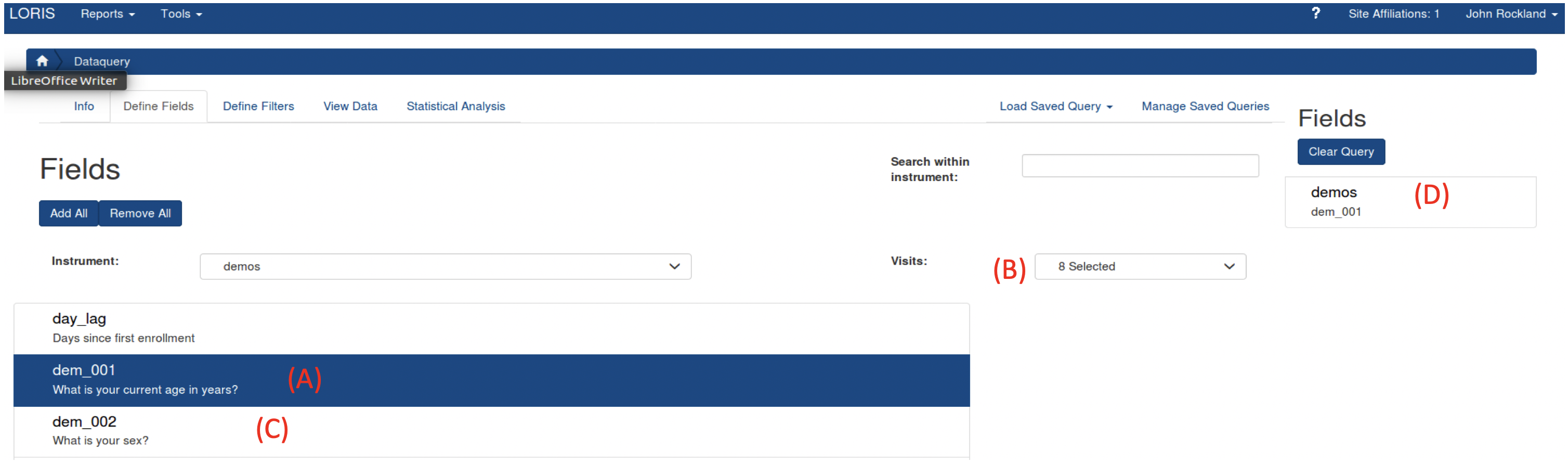
Step 4: Filtering Data
If you choose to filter the data, select the “Define Filters” tab. If you do not want to filter the data, you can skip to Step 4.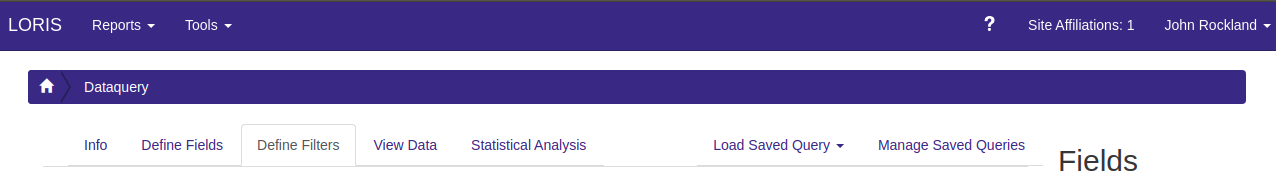
You can use the filter to restrict your data to certain conditions. For example, if you want to restrict the data to only children younger than 10, first select the instrument “demos”, then select demos_01, the “<=” operator, and enter “10”.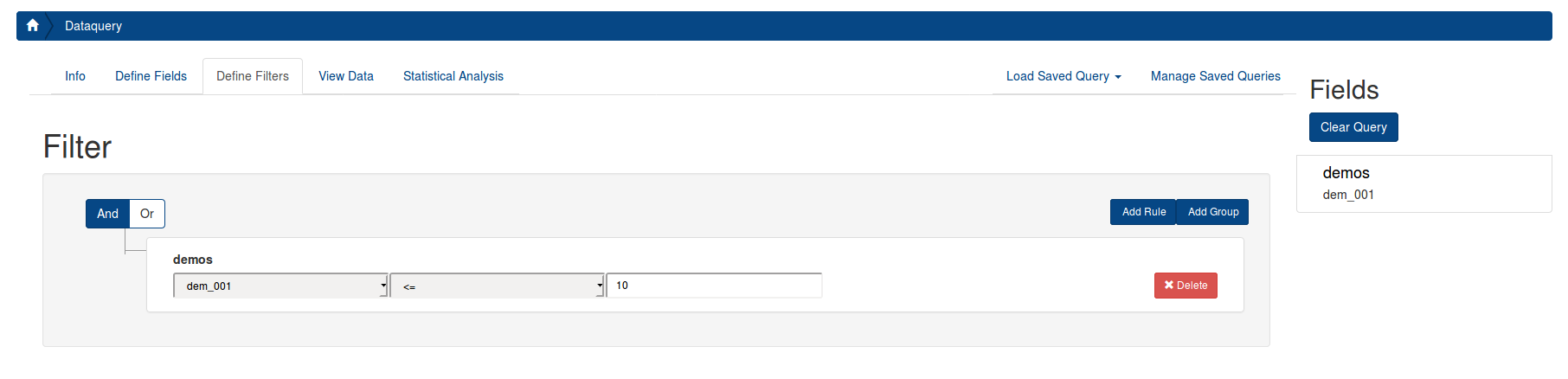
You can also filter on metadata. To filter only participants from the Child Longitudinal Study, you must select the “studies” field within the “demos” instrument, select “contains”, and write the study name or choose from the drop down [options: Longitudinal_Child, Longitudinal_Adult, Discovery, Neurofeedback], as seen below. Please note, if you filter on a variable with missing data, it will not return the complete dataset. The demos instrument is complete for all study participants with any data, so we recommend using it as the choice for your filter. If you want to filter on an instrument of interest and only return participants who have data for that instrument, you can select that instrument for your filter.
Details describing the metadata can be seen here) (Coming soon).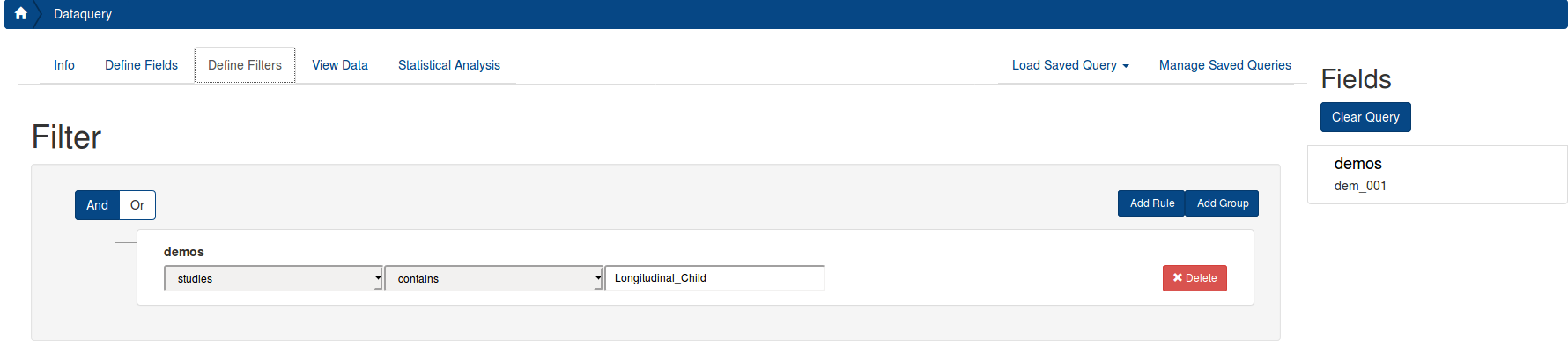
After you have finished defining your filters, you can go to the next step.
Step 5: View and Download Data
Finally to run the query, click the “View Data” tab, and click the “Run Query” button.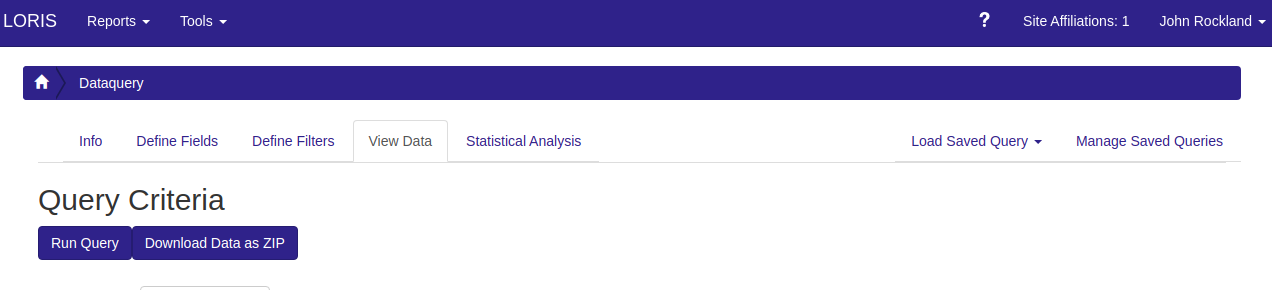
Scroll down and look at the data. Once you are satisfied with the selected and filtered data, you can export the data with the “Download Table as CSV” button.